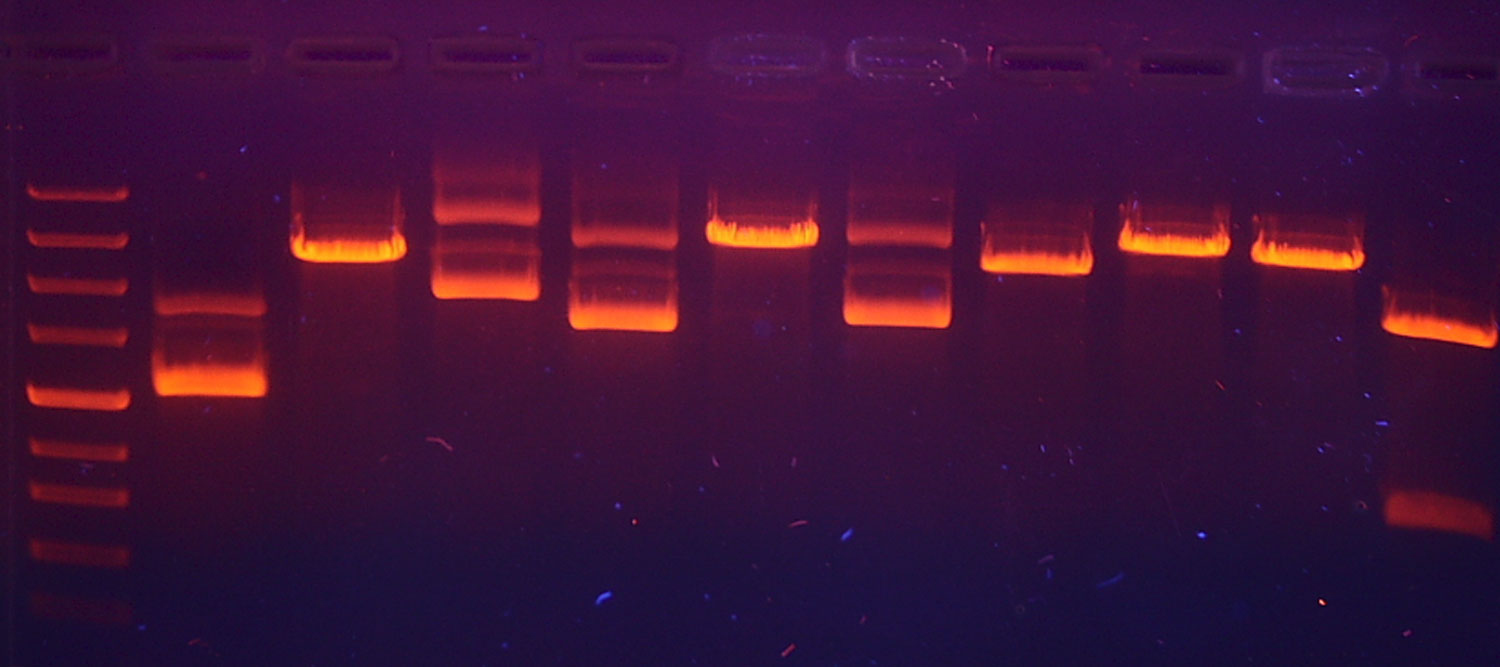
LFTDI student and CIB PhD Candidate, Nidhi Sheth, was awarded an NIJ Graduate Fellowship for her work on “Selectively analyzing and interpreting DNA from multiple donors with a full Single-Cell strategy“. Nidhi is part of a multi-institutional, inter-disciplinary team of computer scientists, applied mathematicians, data scientists and chemists interested in developing a forensically relevant single-cell pipeline aimed at solving the forensic DNA mixture problem.
Forensic Science Center of Excellence Hosts Webinar Featuring LFTDI’s NOCIt Inference Platform
Dr. Grgicak accepts invitation to join the Editorial Board of Electrophoresis
Dr. Grgicak joins the Editorial Board of Electrophoresis, a scientific journal dedicated to topics that include new or improved analytical and preparative methods, sample preparation, development of theory, and innovative applications of electrophoretic and liquid phase separations methods in the study of nucleic acids, proteins, carbohydrates natural products, pharmaceuticals, food analysis, environmental species and other compounds of importance to the life sciences.
LFTDI inventors acquire U.S. Patent on NOCIt technology
Patent US 10,504,614 B2 was issued to Dr. Grgicak & Dr. Lun et al. for their “Systems and Methods for Determining an Unknown Characteristic of a Sample”.
This patent describes the invention underlying NOCIt, the system that allows a full evaluation of the entire forensic DNA signal across all probable number of contributors.
Dr. Grgicak accepts NIST/NIJ invitation to join the Expert Working Group on Human Factors in Forensic DNA Interpretation
The Expert Working Group on Human Factors in Forensic DNA Interpretation is charged with conducting a scientific assessment on the effects of human factors in forensic DNA examination with the goal of recommending approaches to improve its practice and reduce the likelihood of errors. The Working Group will evaluate relevant bodies of scientific literature and technical knowledge to develop its recommendations and will publish a report of its findings.
Dr. Grgicak is honored to be part of this group and looks forward to participating.
Dr. Grgicak joins OSAC’s Biology/DNA Scientific Area Committee
LFTDI Student Nidhi Sheth Awarded NIJ Travel Grant to Present at PITTCON 2020
LFTDI student and CIB PhD candidate, Nidhi Sheth, was awarded an NIJ (National Institute of Justice) Travel Grant to present her work “Deconvolution of Mixture Samples using Single-Cell Techniques for Forensic Human Identification” on March 03 at PITTCON Conference & Expo during the NIJ Poster Session.
LFTDI Presents Developmental Validation of NOCIt at NEAFS
Integrating Validated A Posteriori Probabilities on the Number of Contributors into Forensic Interpretation Pipelines for Full DNA Profile Interpretation will be presented at the 2019 NEAFS conference in Lancaster PA by Dr. Catherine M. Grgicak.
Here she will summarize the developmental validation of NOCIt – a software that renders the a posteriori probability distribution on the number of contributors to complex DNA mixtures – with a large-scale forensically relevant dataset. Notably, Dr. Grgicak shall demonstrate how the APP is used to calculate a full weight-of-evidence (log likelihood ratio or LR) without the need to designate that a particular NOC explains the evidence.
LFTDI Awarded NIST Grant to Develop In Silico MPS Forensic Laboratory
| Dr. Grgicak was awarded a two-year grant from the NIST Measurement Science & Engineering Grant Program to develop an in silico NGS forensic laboratory. |
| The goal is to develop an in silico forensic laboratory using forensically relevant next-generation sequencing data in order to test multifarious laboratory scenarios and optimize processing decisions, improving forensic validation procedures. These in silico data can also be used to train machine learning and deep learning tools that require robust and large training sets, enhancing forensic analysis and interpretation procedures. |