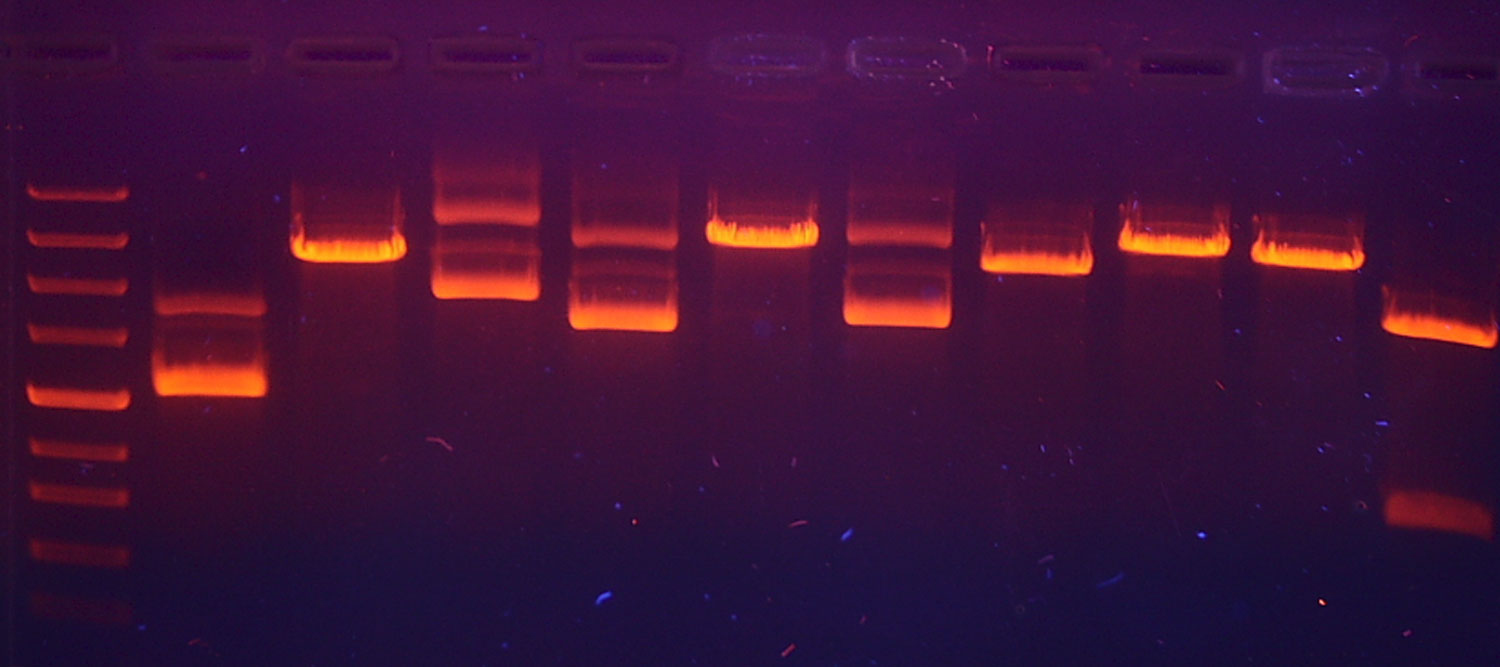
We modify our full in silico DNA pipeline to focus on resolving signal-to-noise within the single-copy regime.
The paper describes system design and ways in which the system can be implemented into forensic DNA validation and optimization.
http://www.sciencedirect.com/science/article/pii/S1872497317301916
Paper Highlights
- •An in silico DNA system is parameterized with the laboratories own experimental data, resulting in predictions of optimal laboratory settings.
- •Noise and allele peak height distributions from a single copy of DNA are used to assess signal-to-noise resolution.
- •Optimal signal detection thresholds, or analytical thresholds, for casework are obtained, if necessary.
- •We demonstrate that metrics of signal quality for simulated and experimental data are consistent.
- •This is a systematic method for evaluating EPG quality and is a critical step towards standardizing the post-PCR process.